Horizontal gene transfer in bacterial biofilms
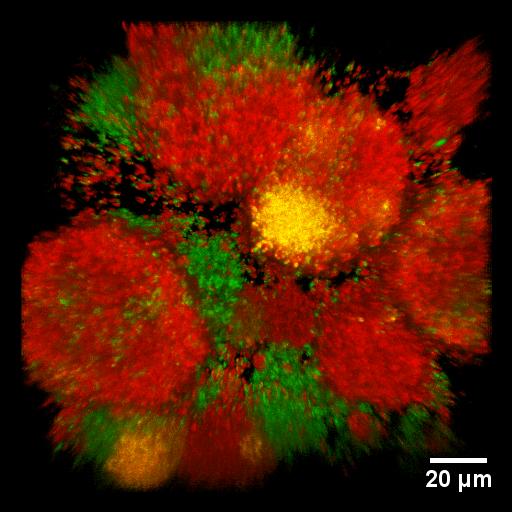
Development of multi-drug resistance in bacterial biofilms. We generated gonococcal strains that have genetically linked antibiotic resistance markers and fluorescent proteins (mcherry combined with resistance against spectinomycin, egfp combined with resistance against erythromycin). In the microscopic image, the color is a marker for drug resistance. By gene transfer, the bacteria can acquire multi-drug resistance, showing up in yellow.
In natural ecosystems microorganisms grow as surface attached, matrix-enclosed communities called biofilms. Biofilms are considered ideal reaction chambers for horizontal gene transfer and development of multi-drug resistance. The rate at which genes are exchanged within biofilms and the factors that govern the exchange rate are unknown. We are quantifying the acquisition of double-drug resistance by gene transfer between bacteria with single resistances.
Focusing on double-resistant transformants in range expansion assays using fluorescence microscopy, we analyze population dynamics and horizontal gene transfer. A continuous-flow system that allows for the non-invasive real-time monitoring of biofilm structure combined with confocal laser microscopy is used to visualize the acquisition of multi-drug resistance by gene transfer. The architecture of the biofilm is controlled by soft lithography and surface microstructuring.
We found that intact biofilms enable efficient gene exchange. The spreading of beneficial double-resistant transformants is strongly dependent on biofilms architecture, since bacteria within it exhibit a higher antibiotic tolerance. We propose that while biofilms help generate multi-resistant strains, selection may take place mostly after dispersal from the biofilm.